Achievement
We implement 2 different multitask learning (MTL) techniques, hard parameter sharing and cross-stitch, to train a word-level convolutional neural network (CNN) specifically designed for automatic extraction of cancer data from unstructured text in pathology reports. We show the importance of learning related information extraction (IE) tasks leveraging shared representations across the tasks to achieve state-of-the-art performance in classification accuracy and computational efficiency.
Significance and Impact
MTCNNs offered superior performance across all 5 tasks in terms of classification accuracy as com-pared with the other machine learning models. Based on retrospective evaluation, the hard parameter sharing and cross-stitch MTCNN models correctly classified 59.04% and 57.93% of the pathology reports respectively across all 5 tasks. The baseline models achieved 53.68% (CNN), 46.37% (RFC), and 36.75% (SVM). Based on prospective evaluation, the percentages of correctly classified cases across the 5 tasks were 60.11% (hard parameter sharing), 58.13% (cross-stitch), 51.30% (single-task CNN), 42.07% (RFC), and 35.16% (SVM). Moreover, hard parameter sharing MTCNNs outperformed the other models in computational efficiency by using about the same number of trainable parameters as a single-task CNN.
Research Details
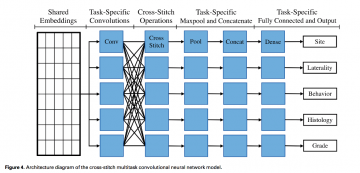
- Multitask CNN (MTCNN) attempts to tackle document information extraction by learning to extract multiple key cancer characteristics simultaneously.
- We trained our MTCNN to perform 5information extraction tasks: (1) primary cancer site (65 classes), (2) laterality (4 classes), (3) behavior (3classes), (4) histological type (63 classes), and (5) histological grade (5 classes).
- We evaluated the performance on a corpus of 95 231 pathology documents (71 223 unique tumors) obtained from the Louisiana Tumor Registry.
- We compared the performance of the MTCNN models against single-task CNN models and 2 traditional machine learning approaches, namely support vector machine (SVM) and random forest classifier (RFC).
Publication
Mohammed Alawad, Shang Gao, John X Qiu, Hong Jun Yoon, J Blair Christian, Lynne Penberthy, Brent Mumphrey, Xiao-Cheng Wu, Linda Coyle, Georgia Tourassi, Automatic extraction of cancer registry reportable information from free-text pathology reports using multitask convolutional neural networks, Journal of the American Medical Informatics Association, Volume 27, Issue 1, January 2020, Pages 89–98, https://doi.org/10.1093/jamia/ocz153
Funding
This work has been supported in part by the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) program established by the U.S. Department of Energy and the National Cancer Institute of the National Institutes of Health. This work was performed under the auspices of the U.S. Department of Energy by Argonne National Laboratory under Contract DE-AC02-06-CH11357, Lawrence Livermore National Laboratory under Contract DEAC52- 07NA27344, Los Alamos National Laboratory under Contract DE-AC5206NA25396, and Oak Ridge National Laboratory under Contract DE-AC05-00OR22725. This research used resources of the Oak Ridge Leadership Computing Facility at the Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05- 00OR22725.
Overview
The hard parameter sharing MTCNN offers superior classification accuracy for automated coding support of pathology documents across a wide range of cancers and multiple information extraction tasks while maintaining similar training and inference time as those of a single task–specific model.
Last Updated: May 28, 2020 - 4:01 pm