Achievement
Generative machine learning models, including GANs (Generative Adversarial Networks), are a powerful tool towards searching chemical space for desired functionalities. Here, we have presented a strategy for promoting search beyond the original training set using incremental updates to the data. Our approach builds upon the concepts of selection and recombination common in Genetic Algorithms and can be seen as a step towards automating the typically manual rules for mutation. Our results suggest that updates to the data enable a larger number of compounds to be explored, leading to an increase in high performing candidates compared to a fixed training set.
Significance and Impact
Generative models in machine learning seek to recreate the distribution underlying a given set of data. After modeling the distribution, new samples can be drawn that extend the original data. One type of generative approach, known as a Generative Adversarial Network (GAN), has been widely used in many applications from image generation to drug discovery. Recent studies have utilized GANs to search the space of possible molecules for drug design, developing models that can generate compounds with a desired feature set.
Although generative models (and GANs) have many advantages for finding new molecules, a key limitation is the propensity for mode collapse. In mode collapse, the model distribution collapses to cover only a few samples from the training data. Beyond mode collapse, it is intuitively expected that a given generative model will be limited by the training data used (i.e., there is no standard way to guide the generative model in areas of parameter space that it has never encountered in training). This limitation hinders the use of GANs in search applications such as drug discovery. To overcome mode collapse, several approaches have been investigated including updating the loss function to promote diversity. However, these approaches rely on comparisons to a fixed training data set, which continues to hinder search applications.
Here, we build upon recent work utilizing GANs for small molecule discovery by introducing a new approach for training. Our approach enables augmented search through incremental updating of the training data set using ideas from Genetic Algorithms. Novel and valid molecules that are generated by the model are stored during a training interval. Then, the training data is updated through a replacement strategy, which can be guided or random. Training resumes and the process is repeated. Our results show that this approach can alleviate the decrease in new molecules generated that occurs for a standard GAN during training. Furthermore, we utilize recombination between generated molecules and the training data to increase new molecule discovery. Introducing replacement and recombination into the training process empowers the use of GANs for broader searches in drug discovery.
Research Details
- Data: The original training data used for all models was taken from QM9, a subset of the GDB-17 chemical database. The data was downloaded from deepchem1 and then processed using rdkit, with any molecules that caused errors during sanitization removed. To modify the training data to include larger molecules, a subset of the ZINC rings data set was used.
- Models: The GAN was implemented using pytorch, with both the discriminator and generator consisting of four fully connected layers. The generator received as input normally distributed random vectors. The output of the generator was an adjacency matrix with the off-diagonal elements specifying the bond order and the on-diagonal elements specifying the atom type. The discriminator received as input the one hot representation of the adjacency matrix and output a single real number.
- New molecules:
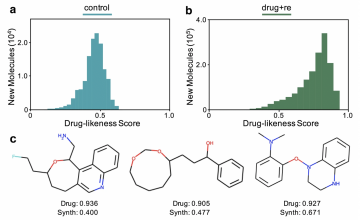
- produced for different replacement strategies: For ‘control’, the training data is fixed. For ‘random’, molecules from the generator randomly replace molecules in the training data. For ‘drug’, molecules from the generator only replace training samples if they have a higher drug-likeness score.
- produced for different replacement strategies with recombination. For ‘control+re’, the training data is fixed. For ‘random+re’, molecules from the generator randomly replace molecules in the training data. For ‘drug+re’, molecules from the generator only replace training samples if they have a higher drug-likeness score.
Citation and DOI:
A. E. Blanchard, C. Stanley and D. Bhowmik; Using GANs with adaptive training data to search for new molecules, J Cheminform (2021) 13:14; DOI: https://doi.org/10.1186/s13321-021-00494-3
Overview
The process of drug discovery involves a search over the space of all possible chemical compounds. Generative Adversarial Networks (GANs) provide a valuable tool towards exploring chemical space and optimizing known compounds for a desired functionality. Standard approaches to training GANs, however, can result in mode collapse, in which the generator primarily produces samples closely related to a small subset of the training data. In contrast, the search for novel compounds necessitates exploration beyond the original data. Here, we present an approach to training GANs that promotes incremental exploration and limits the impacts of mode collapse using concepts from Genetic Algorithms.
Last Updated: April 19, 2021 - 11:29 am